Generation 3 Library Platform
Generation 3 Library Platform

Discovery Sciences
Protein Sciences
Antibody Libraries
Generation 3 Platform
Discovery Campaigns
Library Platform Transfers
Affinity Maturation
AbXtract
Our Terms
In Vitro Immunology
Additional Specialty Testing Services
Specifica’s Generation 3 Library Platform has quality built in by design. Developability, extremely high diversity, and the exclusion of sequence liabilities are intrinsic to the four sub-libraries making up the platform, each of which is based on a therapeutic antibody scaffold chosen for its biophysical properties, lack of liabilities and germline gene variety.
Diversity is derived from natural CDR sequences: HCDR3s are amplified directly from purified B cells, while replicated natural CDRs —derived from Specifica’s databases— are used for the rest. The same unique HCDR3 diversity can be repackaged into different formats, including VHH, scFv and Fab. In the case of scFv and Fab, libraries can also be made with common light chains.
Drug-Like Antibodies
The COVID-19 pandemic accelerated real-world discovery to an unprecedented pace. It also facilitated comparison of different methods of antibody generation in terms of efficiency, as well as affinity to target, neutralizing potency, and developability.
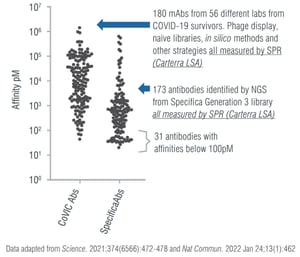 Specifica used this opportunity to analyze our propriety method. Our team developed an antibody panel directly from our naïve library platform and compared it to antibodies from the Coronavirus Immunotherapy Consortium (CoVIC) dataset, which included 180 antibodies from 56 labs worldwide.1
Specifica used this opportunity to analyze our propriety method. Our team developed an antibody panel directly from our naïve library platform and compared it to antibodies from the Coronavirus Immunotherapy Consortium (CoVIC) dataset, which included 180 antibodies from 56 labs worldwide.1
Using the same platform as the Consortium (Carterra LSA), Specifica’s platform generated antibodies with higher affinities and neutralization than those in the CoVIC dataset and comparable to approved antibodies.2
Next-Generation Sequencing (NGS)
We use next-generation sequencing throughout library construction and downstream selection output analyses. This allows us to directly measure the diversity of each exclusive library we build, to ensure it reaches our high standards.
By applying NGS and AbXtract, our in-house bioinformatic analysis platform, to selection outputs, we are usually able to identify one hundred to one thousand different clusters. Once representative cluster sequences have been identified, gene synthesis is used to go “from sequence to clone,” allowing access to the most promising leads.
1 Kathryn M Hastie KM, Li H, Bedinger D, et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study. Science. 2021 Oct 22;374(6566):472-478. doi: 10.1126/science.abh2315.
2 Ferrara F, Erasmus MF, D’Angelo S, et al. A pandemic-enabled comparison of discovery platforms demonstrates a naïve antibody library can match the best immune-sourced antibodies. Nat Commun. 2022 Jan 24;13(1): 462. doi: 10.1038/s41467-021-27799-z.